Diversity: the 'pupula group'
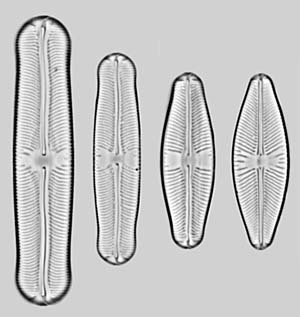
Description
In the 'pupula group', the valves are very
variable in outline,
from linear to elliptical, and have capitate, subcapitate, rostrate,
acute or rounded
poles. All have
polar
bars. The axial
area is narrow, and
the central
area is generally ill-defined and ± rectangular or
bow-tie-shaped, rarely rounded. The axial area and
adjacent parts of the striae may or may not be
depressed below
the rest of the valve face, with or
without a conopeum; if present, the conopea do not
traverse the centre of the valve. The effect in the
LM is to create two
grey longitudinal
areas running parallel to the raphe slits, from
near the centre to the poles (see the right-hand image), or no
areas or lines.
The narrow axial area and ill-defined central areas and
presence of polar bars separate
the
'pupula group' from
the 'americana group'. The presence of polar bars separates
the
'pupula group' from the 'laevissima group'. The ill-defined central
area and absence of conopea from the central part of the
valve separate the
'pupula group' from the 'bacillum group'.
Species list
- Sellaphora
auldreekie D.G. Mann & S.M. McDonald
- Sellaphora baicalopupula Kulikovskiy, Lange-Bertalot & Metzeltin
- Sellaphora bisexualis D.G. Mann & K.M. Evans
- Sellaphora
blackfordensis D.G. Mann & S. Droop
- Sellaphora
californica Potapova
- Sellaphora
capitata D.G. Mann & S.M. McDonald
- Sellaphora caput K.M. Evans & D.G. Mann
- Sellaphora
densistriata (Lange-Bertalot
& Metzeltin) Lange-Bertalot
& Metzeltin
- Sellaphora ellipticolanceolata Metzeltin, Lange-Bertalot & Nergui
- Sellaphora
garciarodriguezii Metzeltin & Lange-Bertalot
- Sellaphora hohnii Potapova & Ponader
- Sellaphora khangalis Metzeltin & Lange-Bertalot
- Sellaphora kusberi Metzeltin, Lange-Bertalot & Nergui
- Sellaphora
lanceolata D.G. Mann & S. Droop
- Sellaphora
lange-bertalotii Metzeltin
- Sellaphora
laterostellata Metzeltin
& Lange-Bertalot
- Sellaphora
laterostrata Metzeltin
& Lange-Bertalot
- Sellaphora
macedonica Levkov & Metzeltin
- Sellaphora
malombensis O. Müller
- Sellaphora
mantasoana Metzeltin
& Lange-Bertalot
- Sellaphora marvanii Poulicková & D.G. Mann
- Sellaphora
mereschkowskii O. Müller
- Sellaphora
mereschkowskii var. recta
O. Müller
- Sellaphora meridionalis Potapova & Ponader
- Sellaphora
mutata (Krasske) Lange-Bertalot
- Sellaphora
mutatoides Lange-Bertalot
& Metzeltin
- Sellaphora
nyassensis O. Müller
- Sellaphora
nyassensis f. minor O.
Müller
- Sellaphora
nyassensis var. capitata
O. Müller
- Sellaphora
nyassensis var. elliptica
O. Müller
- Sellaphora
nyassensis var. longirostris
O. Müller
- Sellaphora
obesa D.G. Mann & M.M. Bayer.
- Sellaphora
omuelleri Metzeltin
& Lange-Bertalot
- Sellaphora
paenepupula Metzeltin
& Lange-Bertalot
- Sellaphora
parapupula Lange-Bertalot
- Sellaphora permutata Metzeltin, Lange-Bertalot & Nergui
- Sellaphora perobesa Metzeltin, Lange-Bertalot & Nergui
- Sellaphora
platycephala O. Müller
- Sellaphora
pseudomutatoides Levokov & Metzeltin
- Sellaphora
pseudopupula (Krasske) Lange-Bertalot
- Sellaphora
pupula (Kützing) Mereschkowski
- Sellaphora
pupula var. densistriata Lange-Bertalot
& Metzeltin
- Sellaphora
pupula var. major
O. Müller
- Sellaphora
pupula var. rectangularis
(W. Gregory) Mereschkowski
- Sellaphora
rectangularis (Gregory) Lange-Bertalot & Metzeltin
- Sellaphora rexii Potapova & Ponader
- Sellaphora
rhombicarea Metzeltin, Lange-Bertalot &
García-Rodríguez
- Sellaphora
rostrata (Hustedt) J.R. Johansen
- Sellaphora
santiagoi Metzeltin, Lange-Bertalot &
García-Rodríguez
- Sellaphora simillima Metzeltin, Lange-Bertalot & Nergui
- Sellaphora
subpupula Levkov & Nakov
- Sellaphora
tapajosensis Metzeltin
& Lange-Bertalot
- Sellaphora
tau (Cleve) Metzeltin
& Lange-Bertalot
- Sellaphora
triundulata Metzeltin
& Lange-Bertalot
- Sellaphora wallacei (Reimer) Potapova & Ponader
- Sellaphora
wummensis J.R. Johansen
Overview of
the
Sellaphora
pupula complex
There are a lot of “S. pupula”
species! The exact number we don’t yet know, but
the more ponds we study, the more we find. We are continually
surprised. For example, our most recent sampling was in
Australia (Victoria) where we chose ponds that at least superficially
resembled Blackford Pond in Edinburgh, i.e. they were urban duck
ponds. When we used a microscope to look at the S. pupulas that
were present we were confident that on the whole, we had sampled a
similar S. pupula
flora to Blackford Pond. We turned out to be right and wrong!
Our current approach to assess diversity is to use a
combination of microscopic and DNA-based techniques. When we
sample a pond or lake, we make a microscope slide of the natural diatom
assemblage. Using a light microscope we work our way through
the slide, looking for S.
pupula. We take lots of photos and for each
distinct S. pupula
we find, we give it a provisional identity. From the same
material we also attempt to collect live Sellaphoras, grow
them up in culture and extract their DNA so that we can also make
DNA-based identifications (“DNA
barcoding”). We routinely use part of a
mitochondrial gene called cytochrome oxidase (or cox1) as an
identification tool. If the cox1 DNA sequence suggests that
we have found a new S.
pupula species, we also obtain chloroplast (rbcL and psaA) and
nuclear (18S and ITS) rDNA sequences.
Using our current approach, we find that we rarely manage to
collect live S. pupula
species that represent all the S.
pupula diversity in a pond or lake. This may be
because some are much less common than others or because the isolation
methods we use are better suited for the survival of some species over
others. We are working on finding better ways to sample
diversity. Despite the present shortcomings, we have still
managed to identify at least 40 different "S. pupula" species.
Even the nature of Sellaphora
diversity is not straightforward. For example, species that
look the most similar are not necessarily the most closely related to
each other; closest relatives are not usually found in the same ponds;
and some of the species that are traditionally recognised as S. pupula are
actually more closely related to S.
bacillum, a diatom whose appearance is very different to S. pupula (Evans et al.
2008). We estimate that S.
pupula has been diversifying for at least 12 million years.
References
Evans, K.M.,
Wortley, A.H. & Mann, D.G. (2007). An
assessment of potential diatom “barcode” genes
(cox1, rbcL, 18S and ITS rDNA) and their effectiveness in determining
relationships in Sellaphora
(Bacillariophyta). Protist 158:
349–364.
Evans, K.M., Wortley, A.H., Simpson, G.E., Chepurnov, V.A.
&
Mann, D.G. (2008, in press). A phylogenetic approach to explore the
nature of
cryptic diversity within the model species complex Sellaphora pupula
agg. (Bacillariophyta). Journal of Phycology


 This site is hosted by the Royal Botanic
Garden Edinburgh.
This site is hosted by the Royal Botanic
Garden Edinburgh.